You can also check this post, written in #blogdown, here: minimal-project-tree-r.
Introduction
The last two days arrived at my twitter feed some discussions on how bad are the following sentences at the beginning of your R script/notebook, sparked by @JennyBryan’s slides at the IASC-ARS/NZSA Conference:
setwd()
and
rm(list = ls())
Jenny Bryan offered a detailed explanation for this, as well as some fixes, in her tidyverse blog post. The main idea was:
- To ensure reproducibility within a stable working directory tree. She proposes the very concise
here::here()but other methods are available such as thetemplateor theProjectTemplatepackages. - To avoid break havoc in other’s computers with
rm(list = ls())!.
All of this buzz around project self-containment and reproducibility motivated me to finish a minimal directory tree that (with some variations) I have been using for this year’s data analysis endeavours.
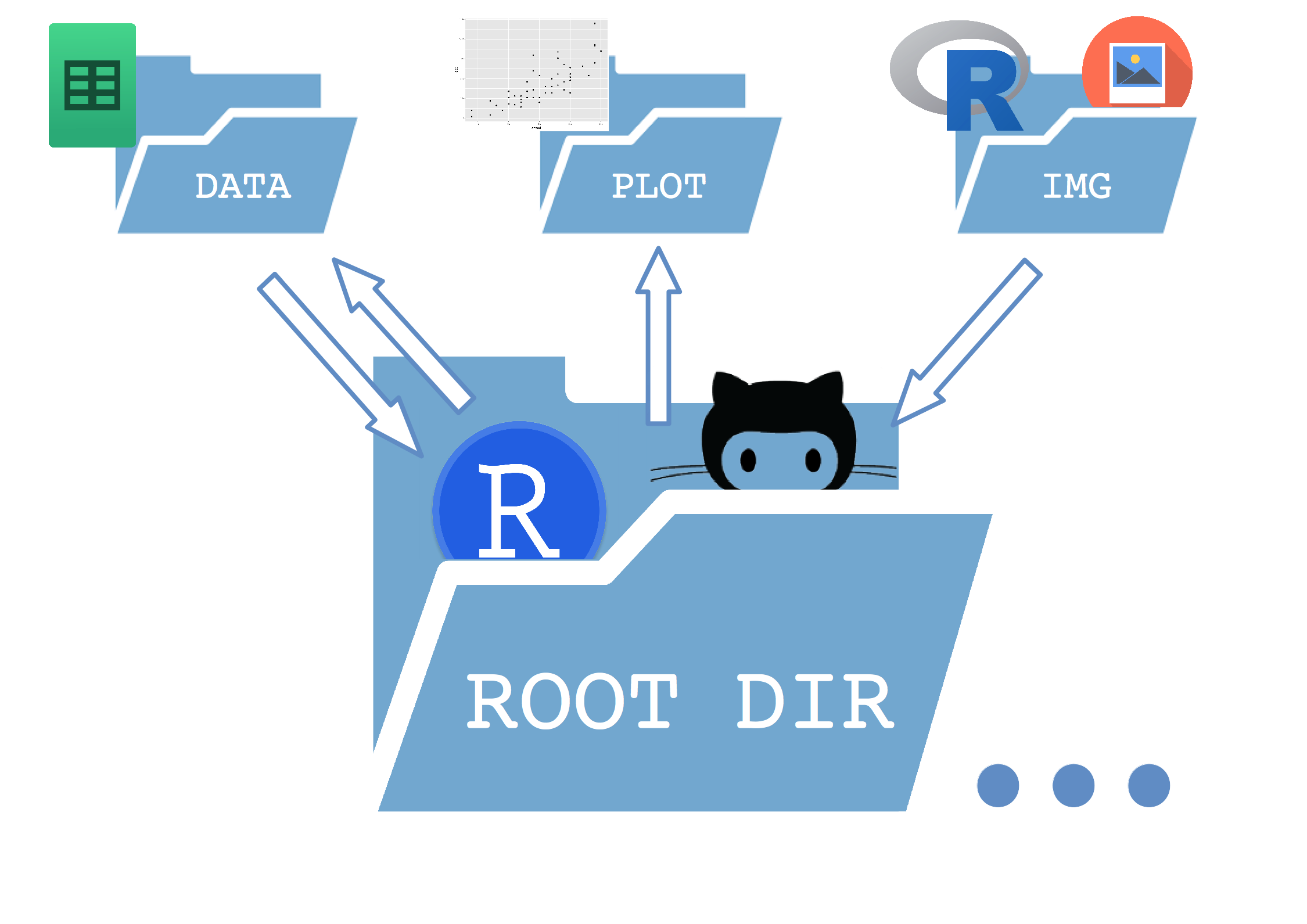
It is a extremely simple tree which separates a /data, a /plot and an /img directory inside the main folder (root)
- The data folder contains both raw data and processed data files saved by R.
- The plot folder contains all the plots saved during the workflow.
- The img folder has every other image (logos, etc) that R takes as an input to build the results.
- Inside the root folder I store the main
.Ror.Rmdscripts.
This ensures that every folder has an unidirectional relationship with the root folder (except the data dir in this case). But the important thing is that the paths in the scripts are set relative to the root folder, so the entire tree can be copied elsewhere and still work as expected.
I also added some more features to the tree:
- An
.Rprojfile - Parametrize the
.Rmdfile - A Git repository so the tree can be conveniently cloned or downloaded, with a .gitignore file:
Here is a sketch of how it works:
 And here is the actual code of the notebook/script. I have not included regular markdown text outside the R chunks, as this template is intended to be changed and filled with new text each time:
And here is the actual code of the notebook/script. I have not included regular markdown text outside the R chunks, as this template is intended to be changed and filled with new text each time:
Script code
# Installs missing libraries on render!
list.of.packages <- c("rmarkdown", "dplyr", "ggplot2", "Rcpp", "knitr", "Hmisc", "readxl")
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) install.packages(new.packages, repos='https://cran.rstudio.com/')library(dplyr)
library(knitr)
library(ggplot2)Working directories
# directory where the notebook is
wdir <- getwd()
# directory where data are imported from & saved to
datadir <- file.path(wdir, "data") # better than datadir <- paste(wdir, "/data", sep="")
# directory where external images are imported from
imgdir <- file.path(wdir, "img")
# directory where plots are saved to
plotdir <- file.path(wdir, "plot")
# the folder immediately above root
Up <- paste("\\", basename(wdir), sep="")
wdirUp <- gsub(Up, "", wdir)
Data import
# Data name (stored as a parameter in the Rmarkdown notebook)
params <- NULL
params$dataname <- "cars"# MSEXCEL
dataname <- params$dataname # archive name
routexl <- paste(datadir, "/", dataname, ".xlsx", sep="") # complete route to archive
library(readxl)
mydata <- read_excel(routexl, sheet = 1) # imports first sheet# CSV / TSV (separated by tabs in this example)
dataname <- params$dataname # archive name
routecsv <- paste(datadir, "/", dataname, ".csv", sep="") # complete route to archive
mydata <- read.csv(paste(routecsv, sep=""),
header = TRUE,
sep = "\t",
dec = ".")Data operations
# Hmisc::describe(mydata)
head(mydata) speed dist
1 4 2
2 4 10
3 7 4
4 7 22
5 8 16
6 9 10p1 <- ggplot(mydata, aes(x=speed, y=dist)) + geom_point()
p1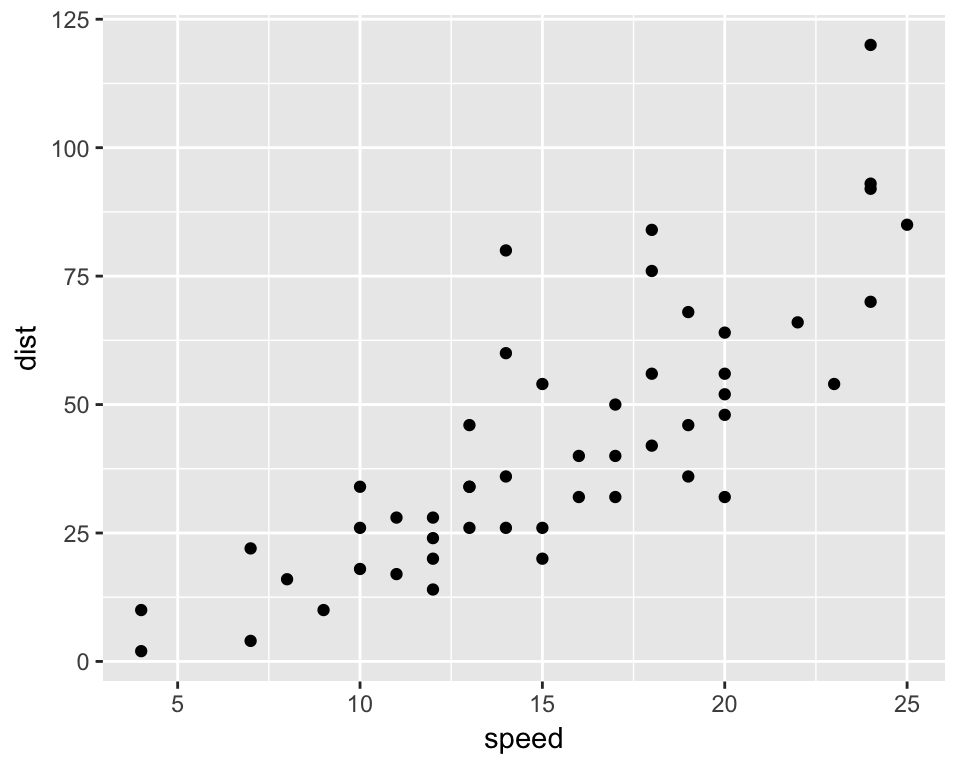
Save plots
# TO PDF
plotname1 <- "p1.pdf"
# TO PNG
plotname2 <- "p1.png"
routeplot1 <- file.path(plotdir, plotname1)
routeplot2 <- file.path(plotdir, plotname2)
ggsave(routeplot1) # (see http://ggplot2.tidyverse.org/reference/ggsave.html)
ggsave(routeplot2) Save data
# RDATA
save(mydata, file="data/mydata.RData")# MSEXCEL # not run
dataname2 <- "mydata" # name we will give to file
routexl2 <- paste(datadir, "/", dataname2, ".xlsx", sep="") # complete route to future archive
library(xlsx)
write.xlsx(mydata, routexl2) # creates archive in specified route# CSV / TSV (separated by tabs in this example)
dataname2 <- "mydata" # name we will give to file
routecsv2 <- paste(datadir, "/", dataname2, ".csv", sep="") # complete route to future archive
write.table(mydata, file = routecsv2, append = FALSE, quote = FALSE, sep = "\t ",
eol = "\n", na = "NA", dec = ".", row.names = FALSE,
col.names = TRUE)This script -and the dir tree that contains it- is saving me a lot of time and headaches (where I’ve put that data?….), I hope it can be also useful for people out there!.
Future improvements
- Add a Makefile?
- Use Travis.ci?
- Etc…

I wonder, what do you think about this? http://projecttemplate.net/getting_started.html
Very interesting, will give it a try!. Have edited the post to add a link to the ProjectTemplate page ^
Nice! I think people underappreciate minimalism when it comes to templates!
Thanks! I wanted to show a very minimal directory tree (tree stump?) so it could be elaborated on by people with no experience organising code. I am teaching R to some students (asynchronously) and need them to really understand how important it is to be tidy with R projects.
They have no idea of coding so I have to give them tools that:
– don’t scare them,
– they can use right away with their data,
– and…. «learn R as you go» 😀
If you use «paste» to construct paths instead of «file.path» somebody should set your computer on fire xD
you’re right! xD no jokes when it comes to PATH, but `paste` worked for me on ms, osx and linux
Thanks for sharing this post. I’ll also suggest adding packrat package in order to maintain the needed packages also under your project tree structure.
Yep! forgot that one! since it is a GitHub repo it will be iterated to add improvements over time, maintaining the tree as simple as possible. Maybe it will be not very useful when it comes to experienced programmers, but some people with no coding background -that I have to teach R to- will learn one important lesson: to be tidy with R projects.
i’ve been following this discussion b/c like most analysts who work on teams i also have to deal with the setwd() nightmare. a lot of projects i work on collaborate in file sharing software like dropbox for example. my dropbox is D:/Dropbox, yours might be C:/Users/…/Dropbox/. things can get nasty with the setwd…
the solution i’ve finally settled on after fighting with this for a long long time is to do the following:
(a) require that every project team member have an initialization file for the project, something like
init-richards-desktop.R
init-elaines-laptop.R
for example here’s what one of my init files looks like:
library(statar)
library(magrittr)
library(dplyr)
library(readr)
library(stringr)
library(reshape2)
library(ggplot2)
library(openxlsx)
# hidden root directories used later
.rtdir <- 'D:/Dropbox/Proj/dNCESSCH'
.rtscratch <- "E:/ExternalDrive/dNCESSCH/scratch"
.rttestscoredata % saveRDS(‘tempdata.rds’) # save temp files
so all intermediate files are saved in some scratch directory. (If you use an init file you can even have this scratch directory be somewhere else completely — for example, not on dropbox. that way you don’t waste your dropbox space. can be a big deal when you’re working with 200gb of data and want to save a sqlite database in scratch…)
then when you’re done with the project — delete the contents of the /scratch directory. you might not see how important it is to be able to do this now, but just wait until you run out of space on your HD and have to meet a deadline in 20 minutes. you’ll be frantic in trying to del something — and be glad you have a few /scratch directories to torpedo
as far as the remove(list=ls()) problem. I’ve never faced a serious problem with people using it. why? because ls() doesn’t show hidden objects. and I put root directories in the global environment as hidden objects (see .rtdir .rtscratch etc). (i don’t want to tell you how long i used r before I realized that you could create objects that remove(list=ls()) won’t torpedo….)
the solution doesn’t require using R projects (I have analysts who work with me who have used R for years and don’t use R projects — also R projects create a storm of files in your dropbox (saving session state every second…), and if you put your projects in shared folders dropbox that can make your teammates pretty angry when they can’t find anything in their dropbox event list. I haven’t figured out a way to stop that storm. so whatcha gonna do), and my solution doesn’t require any package depencies, unlike the here::here() approach.
Things get really messsed up when you have to work in a team and coordinate all the scripts & information, this is a good idea to pick if one ever has to deal with a team of more than only one person! (like me most times). Btw, thanks for sharing the code!
Have you thought of elaborating/writing a blog post about this? It seems like a good idea to me
thanks for the compliment! 🙂
if you find any of these ideas useful as you teach r and work on teams — or if you find better approaches — definitely write what you think up in future blog posts. i’ll be watching for them 🙂
i noticed that some of my comment above got ate because of the less and gtr signs. since @mareviv expressed interest here’s a repost that should be fixed (i just removed the less/gtr signs)
thanks
—–
i’ve been following this discussion b/c like most analysts who work on teams i also have to deal with the setwd() nightmare. a lot of projects i work on collaborate in file sharing software like dropbox for example. my dropbox is D:/Dropbox, yours might be C:/Users/…/Dropbox/. things can get nasty with the setwd…
the solution i’ve finally settled on after fighting with this for a long long time is to do the following:
(a) require that every project team member have an initialization file for the project, something like
init-richards-desktop.R
init-elaines-laptop.R
for example here’s what one of my init files looks like:
library(statar)
library(magrittr)
library(dplyr)
library(readr)
library(stringr)
library(reshape2)
library(ggplot2)
library(openxlsx)
# hidden root directories used later
.rtdir – ‘D:/Dropbox/Proj/dNCESSCH’
.rtscratch – «E:/ExternalDrive/dNCESSCH/scratch»
.rttestscoredata – ‘D:/Dropbox/Proj/dNLSASD’
Sys.setenv(«R_ZIPCMD» = «C:/Users/Richard/Documents/zip300xn-x64/zip300xn-x64/zip.exe») ## path to zip.exe
you see — with an init file I can tell my teammate to set the root directory on his computer custom as a hidden object. i can also have him set other environment variables. in addition i can easily work around the fact that we have large datasets on different directories in dropbox. it seems from my understanding of all this here::here() business, and what you report in this post, that I would have to have all raw data copied into a subdir of each project separately… which can get burdensome when projects depend on other datasets (as they often do in social science research)
all scripts within the project assume that you’ve run your init file first. which means they run assuming that the libraries listed there are attached. i know this COULD cause problems if people want to detach and reattach libraries a lot, but so far it has not really been a problem in my projects (that’s b/c the tidyverse packages all work together pretty well, all thanks goes to the hadley team). this method works really well in RStudio because RStudio KNOWS what libraries you have attached, even if you don’t library() them at the beginning of your script, so autocomplete works and so forth. (btw it doesn’t work well in Spyder because Spyder doesn’t know what imports you have done, so spyder puts squiggly lines everywhere if you use init files and separate your imports from your code. well, it’s not that big of a deal, i can deal with squiggly lines…)
(b) add to the directories you have mentioned above, a /scratch directory
you write: «The data folder contains both raw data and processed data files saved by R.»
i would disagree with that if you plan on working on a project that has any reasonably sized data. why? because I want to minimize the amount of mental energy you use. what I do instead is like
setwd(.rtdir)
setwd(‘data’)
# load data, do stuff with data
setwd(.rtscratch)
df %% saveRDS(‘tempdata.rds’) # save temp files
so all intermediate files are saved in some scratch directory. (If you use an init file you can even have this scratch directory be somewhere else completely — for example, not on dropbox. that way you don’t waste your dropbox space. can be a big deal when you’re working with 200gb of data and want to save a sqlite database in scratch…)
then when you’re done with the project — delete the contents of the /scratch directory. you might not see how important it is to be able to do this now, but just wait until you run out of space on your HD and have to meet a deadline in 20 minutes. you’ll be frantic in trying to del something — and be glad you have a few /scratch directories to torpedo
as far as the remove(list=ls()) problem. I’ve never faced a serious problem with people using it. why? because ls() doesn’t show hidden objects. and I put root directories in the global environment as hidden objects (see .rtdir .rtscratch etc). (i don’t want to tell you how long i used r before I realized that you could create objects that remove(list=ls()) won’t torpedo….)
the solution doesn’t require using R projects (I have analysts who work with me who have used R for years and don’t use R projects — also R projects create a storm of files in your dropbox (saving session state every second…), and if you put your projects in shared folders dropbox that can make your teammates pretty angry when they can’t find anything in their dropbox event list. I haven’t figured out a way to stop that storm. so whatcha gonna do), and my solution doesn’t require any package depencies, unlike the here::here() approach.
Pingback: 7 étapes pour organiser son travail sous R – DellaData